Modeling Related Themes
Back to research thme listOverview
With the progress of research over the past 20 or 30 years, many people believe that the theory of pharmacokinetics or model analysis has been consolidated into the physiological-based pharmacokinetics (PBPK) and that there is no more room for modification of the research techniques. Is this really so? Through my (Hisaka's) own research as well as my experience in developing guidelines for drug interactions and PBPK, I feels that the PBPK model is not a panacea and that its accuracy still needs much improvement. In addition, new technologies, such as iPS cells, have been introduced to wet research in pharmacokinetics, but model analysis has not been able to take full advantage of them. Furthermore, when we look beyond pharmacokinetics to the analysis of drug efficacy and safety using big data, it is still unclear what kind of analysis is applicable. We believe that the field of model research is rather in need of innovation. My research group is therefore developing the following research as of 2021.
Integrated Analysis of Drug Absorption Process (Hisaka, Sato)
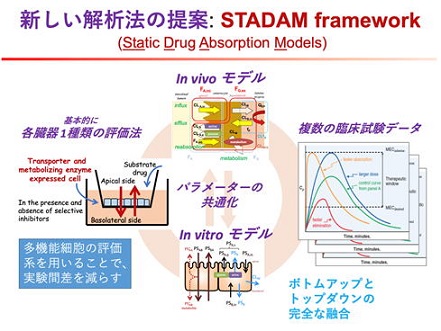
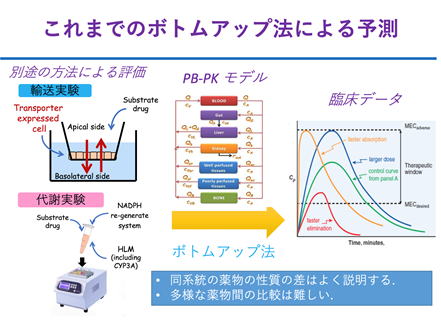
Drug absorption is one of the most difficult processes in ADME to predict because of its variability. Conventional methods based on the PBPK model require a lot of information, and if there is any unclear information during the bottom-up process of building up from in vitro to in vivo, the analysis becomes unreliable. To overcome this shortcoming, we propose a new framework for the analysis of drug absorption processes, which we call "STADAM".
(1) All data, including in vitro and in vivo information, can be analyzed simultaneously and in a completely equivalent manner. This prevents bias due to errors in specific experimental systems or information sources, and enables objective analysis without preconceptions.
(2) Simultaneous analysis of information on a large number of drugs. This allows for appropriate evaluation of properties common to all drugs, and also for appropriate analysis of missing information for some drugs.
(3) In vitro experiments so far have measured only specific activities, but in vivo phenomena are the result of the interaction of many activities. Therefore, in the past, many in vitro experiments were combined and analyzed using the PBPK model and other methods. However, with the recent development of iPS cells, in vitro experimental systems in which multiple activities are expressed simultaneously are already available. STADAM utilizes model analysis to simultaneously evaluate multiple activities in a single in vitro experiment, and in principle, achieves highly accurate predictions that are not affected by inter-experimental variability.
(4) In the small intestine, drug metabolism and transport of P-glycoprotein to the luminal side acts as a barrier to the absorption of foreign substances. STADAM clearly distinguishes between these contributions and effectively uses them to predict absorption.
Research on Drug Interactions (Hisaka, Sato)
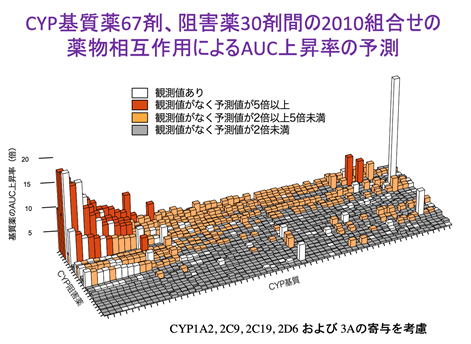
We proposed a new method called "PISCS" to systematically predict and alert drug interactions involving drug-metabolizing enzymes CYP (Hisaka A et al. Clin Pharmacokinet, 2009;48:653), and this concept was incorporated as one of the basic ideas in the "Drug Interaction Guidelines for Drug Development and Appropriate Information Provision" formulated by PMDA in 2018. PISCS continues to evolve and is able to predict blood concentrations of more than 2,000 drug combinations, as shown below. As the technology for predicting drug-drug interactions has improved, it has become clear that the accuracy of the underlying in vitro experiments remains a challenge. We are also working to improve the accuracy of in vitro experiments using cells, microsomes, animals, etc., and using highly sensitive LC-MS/MS measurements.
Various Computer Analysis Techniques (Hisaka)
The research in the CPP is supported by a wide variety of advanced computer analysis techniques, which is exceptional for a pharmacy school. In general model analysis, parameter optimization using nonlinear least squares and numerical computation of simultaneous differential equations are commonly used, and in clinical pharmacokinetics, analysis techniques such as population pharmacokinetics are commonly used. The MCMC method, machine learning techniques such as random forests, gradient-boosted decision trees, and perceptrons, all of which can be run in R, SAS, MATLAB, NONMEM, WinBUGS, Stan, Python, and other environments in CPP. Napp, developed by Hisaka, has long been available as a pharmacokinetic analysis software and is used by many researchers (Hisaka. Pharmacology, 2011;71:168). Since we are experts in modeling, we try to use techniques where the mathematical properties of the model are clear. The MCMC methods are applied to study racial differences in interactions and kinetics, while machine learning is applied to analyze large-scale clinical trials and the effects of diet on the absorption of various drugs, taking their characteristics into account.
Study on Long-term Progression of Chronic Diseases by SReFT (Hisaka)
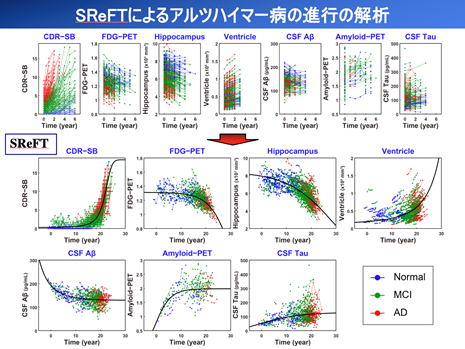
Chronic diseases often last for 20 to 30 years and account for the majority of diseases in the elderly. However, clinical studies are limited to a few years at most, and it is difficult to capture the entirety of chronic diseases. We have developed a completely new analysis technique called SReFT and applied it to the analysis of Alzheimer's disease in collaboration with a research group at the University of Tokyo (Ishida T et al. Pharmacol Ther. 2019;105:436). In CPP, SReFT has been applied to the analysis of Parkinson's disease and COPD for further research. SReFT is a significant extension of the population pharmacokinetic analysis method. In addition to estimating long-term changes in biomarkers with disease progression, SReFT can analyze the risk of influencing these changes. We expect that SReFT analysis will provide important clues to questions such as the causes of chronic diseases and which stage of the disease patients should be treated.
Model Analysis of Disease and Drug Effects (Hisaka, Sato)
Compared to the analysis of pharmacokinetics, computer modeling of diseases and drug effects is still not so common. In addition to the aforementioned SReFT, the Laboratory of Clinical Pharmacology is engaged in modeling analysis of various diseases and their treatment, such as chronic heart failure, anticoagulants, diabetes, cancer, and infectious diseases. In addition to literature survey-based meta-analyses of model bases (MBMA), various information sources are used for these studies, such as the public disclosure system of individual subject information of clinical trials and Japanese and US databases of adverse drug reactions. Currently, working graduate students in clinical development from several companies are enrolled in CPP, enabling us to analyze clinical trial information from a professional perspective, which is rare in academia.